Classification on Titanic Data set¶
%pylab inline
import pandas as pd
matplotlib.rcParams['figure.figsize'] = (10,6)
Populating the interactive namespace from numpy and matplotlib
/home/cokelaer/anaconda2/envs/py35/lib/python3.5/site-packages/IPython/core/magics/pylab.py:161: UserWarning: pylab import has clobbered these variables: ['test', 'clf', 'shuffle'] `%matplotlib` prevents importing * from pylab and numpy "\n`%matplotlib` prevents importing * from pylab and numpy"
The data¶
training= pd.read_csv("data/titanic_training.csv", index_col=0)
test = pd.read_csv("data/titanic_test.csv", index_col=0)
Data description¶
training.head(3)
| pclass | survived | name | sex | age | sibsp | parch | ticket | fare | cabin | embarked | boat | body | home.dest | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 584 | 2 | 1 | Webber, Miss. Susan | female | 32.5 | 0 | 0 | 27267 | 13.00 | E101 | S | 12 | NaN | England / Hartford, CT |
| 328 | 2 | 0 | Angle, Mr. William A | male | 34.0 | 1 | 0 | 226875 | 26.00 | NaN | S | NaN | NaN | Warwick, England |
| 780 | 3 | 1 | Drapkin, Miss. Jennie | female | 23.0 | 0 | 0 | SOTON/OQ 392083 | 8.05 | NaN | S | NaN | NaN | London New York, NY |
Survived : 1 means survived (remember True is 1)
Data visualisation¶
training.survived.value_counts().plot(kind="bar")
title("Survival (1=survived)")
grid()
training[["survived", "age"]].boxplot(by="survived")
<matplotlib.axes._subplots.AxesSubplot at 0x7f8d7ff2bbe0>
training.pclass.value_counts().plot.barh()
title("class distribution")
grid()
Cleanup¶
- Remove useless features or features with lots of missing values
- drop remaining rows with at least one missing values
# many missing values
print(len(training), len(test))
ignore = ["cabin", "home.dest", "body", "embarked", "boat"]
training.drop(ignore, axis=1, inplace=True)
test.drop(ignore, axis=1, inplace=True)
training.dropna(inplace=True)
test.dropna(inplace=True)
print(len(training), len(test))
916 393 726 319
Let us replace male by 0 and female by 1
training = training.replace("male", 0).replace("female", 1)
test = test.replace("male", 0).replace("female", 1)
Find a relevant feature of interest¶
male_survived = len(training.query("sex==0 and survived==1"))
N_male = len(training.query("sex==0"))
male_survival_rate = male_survived / N_male * 100
female_survived = len(training.query("sex==1 and survived==1"))
N_female = len(training.query("sex==1"))
female_survival_rate = female_survived / N_female * 100
bar([0,1], [male_survival_rate, female_survival_rate ])
xticks([0,1], ['male', 'female'])
xlabel("survival rate (%)")
<matplotlib.text.Text at 0x7f8d7ff57be0>
The training and test data¶
features = ["sex"]
Y = training.survived.values
X = training.loc[:, features].values
Ytest = test.survived.values
Xtest = test.loc[:,features]
Your own naive classifier¶
Let us predict the survival as follows:
- if it is a woman: survives
- if it is a man: does not survive
This is a simple as :
Y_pred = (Xtest.sex == 1)
Accuracy = fraction of all instance that are correctly categorized
sum(Y_pred == Ytest)/len(Y_pred)
0.76802507836990597
What about a proper classifier from sklearn ?¶
from sklearn.linear_model import SGDClassifier
clf = SGDClassifier()
clf.fit(X, Y)
# Let us save the prediction for later
Y_pred_1 = clf.predict(Xtest)
clf.score(Xtest, Ytest)
0.76802507836990597
The score is not deterministic ! See later
Stochastic Gradient Descent classifier¶
Stochastic Gradient Descent (SGD) is a simple yet very efficient approach to discriminative learning of linear classifiers under convex loss functions such as (linear) Support Vector Machines and Logistic Regression.
SGD has been successfully applied to large-scale and sparse machine learning problems often encountered in text classification and natural language processing.
The advantages of Stochastic Gradient Descent are:
- Efficiency.
- Ease of implementation (lots of opportunities for code tuning).
The disadvantages of Stochastic Gradient Descent include:
- SGD requires a number of hyperparameters such as the regularization parameter and the number of iterations.
- SGD is sensitive to feature scaling.
Binary classifiers¶
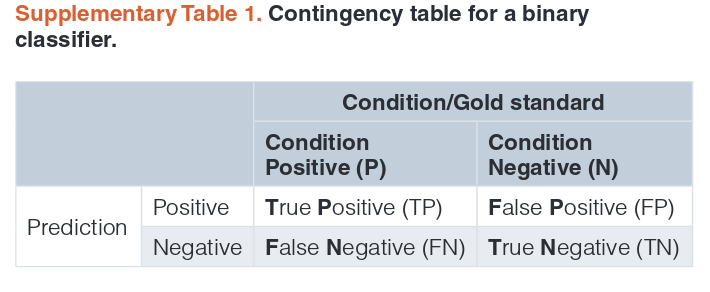
See page 17-20 from https://f1000research.com/articles/4-1030/v1 for all definitions
ROC curve¶
from sklearn import metrics
fpr1, tpr1, thresholds = metrics.roc_curve(Ytest, Y_pred_1, pos_label=1)
metrics.roc_auc_score(Ytest, Y_pred_1)
0.75437962070073927
plot(fpr1, tpr1, "o-")
plot([0,1], [0,1], "-k")
[<matplotlib.lines.Line2D at 0x7f8d7ff55630>]
scores = []
for this in range(20):
clf = SGDClassifier()
clf.fit(X, Y)
scores.append(clf.score(Xtest, Ytest))
prediction = clf.predict(Xtest)
fpr, tpr, thresholds = metrics.roc_curve(Ytest, prediction, pos_label=1)
plot(fpr, tpr, "-o")
mean(scores), std(scores)
(0.6608150470219436, 0.13932608845914329)
Add more features¶
features = ["sex", "pclass"]
Y = training.survived.values
X = training.loc[:, features].values
Ytest = test.survived.values
Xtest = test.loc[:,features].values
subplot(2,2,1)
training.query("sex==0 and pclass==3").survived.value_counts().plot(
kind="bar", label='male class2')
legend(); xticks([0,1], ["survived", "died"])
subplot(2,2,2)
training.query("sex==0 and pclass!=3").survived.value_counts().plot(
kind="bar", label='male class 2/3')
legend(); xticks([0,1], ["survived", "died"])
subplot(2,2,3)
training.query("sex==1 and pclass==3").survived.value_counts().plot(
kind="bar", label='female class3')
legend(); xticks([0,1], ["survived", "died"])
subplot(2,2,4)
training.query("sex==1 and pclass!=3").survived.value_counts().plot(
kind="bar", label='female class 2/3')
legend(); _ = xticks([0,1], ["survived", "died"])
clf = SGDClassifier()
clf.fit(X, Y)
clf.score(Xtest, Ytest)
0.72413793103448276
Note that it is stochastic so sometimes 2 features is worst than 1 but on average it is better
scores = []
for this in range(20):
clf = SGDClassifier().fit(X, Y)
scores.append(clf.score(Xtest, Ytest))
prediction = clf.predict(Xtest)
fpr, tpr, thresholds = metrics.roc_curve(Ytest, prediction, pos_label=1)
plot(fpr, tpr, "-o")
mean(scores), std(scores)
(0.6967084639498432, 0.10295729883404001)
What about KNearestNeigbors classifier ?¶
features = ["sex"]
Y = training.survived.values
X = training.loc[:, features].values
Ytest = test.survived.values
Xtest = test.loc[:,features].values
from sklearn.neighbors import KNeighborsClassifier
clf = KNeighborsClassifier()
clf.fit(X, Y)
clf.score(Xtest, Ytest)
0.76802507836990597
features = ["sex", "pclass",]
Y = training.survived.values
X = training.loc[:, features].values
Ytest = test.survived.values
Xtest = test.loc[:,features].values
from sklearn.neighbors import KNeighborsClassifier
clf = KNeighborsClassifier()
clf.fit(X, Y)
clf.score(Xtest, Ytest)
0.72413793103448276
Decision Trees¶
Decision Trees (DTs) are a non-parametric supervised learning method used for classification and regression. The goal is to create a model that predicts the value of a target variable by learning simple decision rules inferred from the data features.
some advantages:
- simple to understand (see later)
- low cost (log)
- handle categorical and numerical data (less data preparation)
some drawbacks:
- can be unstable. small variation may lead to different results
- may perform overfitting (over complex tree)
from sklearn import tree
clf = tree.DecisionTreeClassifier()
clf.fit(X, Y)
clf.score(Xtest, Ytest)
0.78056426332288398
import pydotplus
dot_data = tree.export_graphviz(clf, out_file=None)
graph = pydotplus.graph_from_dot_data(dot_data)
dot_data = tree.export_graphviz(clf, out_file=None,
feature_names=features,
class_names=["survived", "died"],
filled=True, rounded=True, special_characters=True)
graph = pydotplus.graph_from_dot_data(dot_data)
from IPython.display import Image
Image(graph.create_png())
RandomForest¶
Random forests are an ensemble learning method for classification (and regression) that operate by constructing a multitude of decision trees at training time and outputting the class that is the mode of the classes output by individual trees.
-- wikipedia
From sklearn website:
In random forests each tree in the ensemble is built from a sample drawn with replacement (i.e., a bootstrap sample) from the training set.
In addition, when splitting a node during the construction of the tree, the split that is chosen is no longer the best split among all features.
Instead, the split that is picked is the best split among a random subset of the features. As a result of this randomness, the bias of the forest usually slightly increases (with respect to the bias of a single non-random tree) but, due to averaging, its variance also decreases, usually more than compensating for the increase in bias, hence yielding an overall better model.
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators=5)
clf.fit(X, Y)
clf.score(Xtest, Ytest)
0.78056426332288398
Neural network¶
from sklearn.neural_network import MLPClassifier
clf = MLPClassifier(solver='lbfgs', alpha=1e-5,
hidden_layer_sizes=(10, 1), random_state=1)
_ = clf.fit(X, Y)
clf.score(Xtest, Ytest)
0.78056426332288398
Cross validation for binary classifiers¶
training.head(3)
| pclass | survived | name | sex | age | sibsp | parch | ticket | fare | |
|---|---|---|---|---|---|---|---|---|---|
| 584 | 2 | 1 | Webber, Miss. Susan | 1 | 32.5 | 0 | 0 | 27267 | 13.00 |
| 328 | 2 | 0 | Angle, Mr. William A | 0 | 34.0 | 1 | 0 | 226875 | 26.00 |
| 780 | 3 | 1 | Drapkin, Miss. Jennie | 1 | 23.0 | 0 | 0 | SOTON/OQ 392083 | 8.05 |
features = ["sex", "class"]
Y = training.survived.values
X = training.loc[:, features].values
Ytest = test.survived.values
Xtest = test.loc[:,features].values
from sklearn.model_selection import cross_val_score
clf1 = tree.DecisionTreeClassifier()
clf2 = SGDClassifier()
clf3 = KNeighborsClassifier()
clf4 = RandomForestClassifier(n_estimators=10)
scores1 = cross_val_score(clf1, X, Y, cv=5)
scores2 = cross_val_score(clf2, X, Y, cv=5)
scores3 = cross_val_score(clf3, X, Y, cv=5)
scores4 = cross_val_score(clf4, X, Y, cv=5)
yerr = [std(this) for this in [scores1, scores2, scores3, scores4]]
mus = [mean(this) for this in [scores1, scores2, scores3, scores4]]
errorbar([0,1,2,3], mus, yerr=yerr, xerr=0.1, fmt="o")
ylim([0.5,1])
_ = xticks([0,1,2,3], ["tree", "sgd", "kn", "RF"], rotation=0, fontsize=40)
print(mus)
[0.78376003778932457, 0.58789796882380718, 0.78376003778932457, 0.78376003778932457]
Summary¶
- Select the relevant features
Remove or impute missing data sets
choose a classifier. We have seen:
- KNeighbor - DecisionTree - StochasticGradientbut there are many more. See sklearn website
This section was used to create the training and test set from the whole data¶
from sklearn.utils import shuffle
df = pd.read_csv("data/titanic3.csv")
df = shuffle(df)
training = df.ix[df.index[0:916]]
test = df.ix[df.index[916:]]
training.to_csv("data/titanic_training.csv")
test.to_csv("data/titanic_test.csv")
pd.read_csv("data/titanic_training.csv", index_col=0)
| pclass | survived | name | sex | age | sibsp | parch | ticket | fare | cabin | embarked | boat | body | home.dest | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 584 | 2 | 1 | Webber, Miss. Susan | female | 32.5 | 0 | 0 | 27267 | 13.0000 | E101 | S | 12 | NaN | England / Hartford, CT |
| 328 | 2 | 0 | Angle, Mr. William A | male | 34.0 | 1 | 0 | 226875 | 26.0000 | NaN | S | NaN | NaN | Warwick, England |
| 780 | 3 | 1 | Drapkin, Miss. Jennie | female | 23.0 | 0 | 0 | SOTON/OQ 392083 | 8.0500 | NaN | S | NaN | NaN | London New York, NY |
| 495 | 2 | 0 | Mangiavacchi, Mr. Serafino Emilio | male | NaN | 0 | 0 | SC/A.3 2861 | 15.5792 | NaN | C | NaN | NaN | New York, NY |
| 659 | 3 | 1 | Baclini, Miss. Marie Catherine | female | 5.0 | 2 | 1 | 2666 | 19.2583 | NaN | C | C | NaN | Syria New York, NY |
| 1152 | 3 | 0 | Robins, Mr. Alexander A | male | 50.0 | 1 | 0 | A/5. 3337 | 14.5000 | NaN | S | NaN | 119.0 | NaN |
| 1071 | 3 | 1 | O'Brien, Mrs. Thomas (Johanna "Hannah" Godfrey) | female | NaN | 1 | 0 | 370365 | 15.5000 | NaN | Q | NaN | NaN | NaN |
| 232 | 1 | 0 | Porter, Mr. Walter Chamberlain | male | 47.0 | 0 | 0 | 110465 | 52.0000 | C110 | S | NaN | 207.0 | Worcester, MA |
| 1302 | 3 | 0 | Yousif, Mr. Wazli | male | NaN | 0 | 0 | 2647 | 7.2250 | NaN | C | NaN | NaN | NaN |
| 491 | 2 | 0 | Malachard, Mr. Noel | male | NaN | 0 | 0 | 237735 | 15.0458 | D | C | NaN | NaN | Paris |
| 1275 | 3 | 0 | Vander Planke, Mr. Leo Edmondus | male | 16.0 | 2 | 0 | 345764 | 18.0000 | NaN | S | NaN | NaN | NaN |
| 431 | 2 | 0 | Harper, Rev. John | male | 28.0 | 0 | 1 | 248727 | 33.0000 | NaN | S | NaN | NaN | Denmark Hill, Surrey / Chicago |
| 1121 | 3 | 1 | Peter, Master. Michael J | male | NaN | 1 | 1 | 2668 | 22.3583 | NaN | C | C | NaN | NaN |
| 433 | 2 | 0 | Harris, Mr. Walter | male | 30.0 | 0 | 0 | W/C 14208 | 10.5000 | NaN | S | NaN | NaN | Walthamstow, England |
| 123 | 1 | 1 | Frolicher-Stehli, Mr. Maxmillian | male | 60.0 | 1 | 1 | 13567 | 79.2000 | B41 | C | 5 | NaN | Zurich, Switzerland |
| 414 | 2 | 0 | Gale, Mr. Shadrach | male | 34.0 | 1 | 0 | 28664 | 21.0000 | NaN | S | NaN | NaN | Cornwall / Clear Creek, CO |
| 1077 | 3 | 1 | O'Driscoll, Miss. Bridget | female | NaN | 0 | 0 | 14311 | 7.7500 | NaN | Q | D | NaN | NaN |
| 1158 | 3 | 0 | Rosblom, Mrs. Viktor (Helena Wilhelmina) | female | 41.0 | 0 | 2 | 370129 | 20.2125 | NaN | S | NaN | NaN | NaN |
| 927 | 3 | 0 | Khalil, Mr. Betros | male | NaN | 1 | 0 | 2660 | 14.4542 | NaN | C | NaN | NaN | NaN |
| 1237 | 3 | 0 | Svensson, Mr. Olof | male | 24.0 | 0 | 0 | 350035 | 7.7958 | NaN | S | NaN | NaN | NaN |
| 247 | 1 | 1 | Rothschild, Mrs. Martin (Elizabeth L. Barrett) | female | 54.0 | 1 | 0 | PC 17603 | 59.4000 | NaN | C | 6 | NaN | New York, NY |
| 510 | 2 | 0 | Mudd, Mr. Thomas Charles | male | 16.0 | 0 | 0 | S.O./P.P. 3 | 10.5000 | NaN | S | NaN | NaN | Halesworth, England |
| 191 | 1 | 0 | Loring, Mr. Joseph Holland | male | 30.0 | 0 | 0 | 113801 | 45.5000 | NaN | S | NaN | NaN | London / New York, NY |
| 1210 | 3 | 0 | Skoog, Mr. Wilhelm | male | 40.0 | 1 | 4 | 347088 | 27.9000 | NaN | S | NaN | NaN | NaN |
| 681 | 3 | 0 | Boulos, Mrs. Joseph (Sultana) | female | NaN | 0 | 2 | 2678 | 15.2458 | NaN | C | NaN | NaN | Syria Kent, ON |
| 1195 | 3 | 0 | Shaughnessy, Mr. Patrick | male | NaN | 0 | 0 | 370374 | 7.7500 | NaN | Q | NaN | NaN | NaN |
| 216 | 1 | 1 | Newsom, Miss. Helen Monypeny | female | 19.0 | 0 | 2 | 11752 | 26.2833 | D47 | S | 5 | NaN | New York, NY |
| 644 | 3 | 0 | Asplund, Mr. Carl Oscar Vilhelm Gustafsson | male | 40.0 | 1 | 5 | 347077 | 31.3875 | NaN | S | NaN | 142.0 | Sweden Worcester, MA |
| 856 | 3 | 1 | Healy, Miss. Hanora "Nora" | female | NaN | 0 | 0 | 370375 | 7.7500 | NaN | Q | 16 | NaN | NaN |
| 528 | 2 | 0 | Parkes, Mr. Francis "Frank" | male | NaN | 0 | 0 | 239853 | 0.0000 | NaN | S | NaN | NaN | Belfast |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 264 | 1 | 1 | Simonius-Blumer, Col. Oberst Alfons | male | 56.0 | 0 | 0 | 13213 | 35.5000 | A26 | C | 3 | NaN | Basel, Switzerland |
| 1138 | 3 | 0 | Reed, Mr. James George | male | NaN | 0 | 0 | 362316 | 7.2500 | NaN | S | NaN | NaN | NaN |
| 309 | 1 | 1 | Wick, Miss. Mary Natalie | female | 31.0 | 0 | 2 | 36928 | 164.8667 | C7 | S | 8 | NaN | Youngstown, OH |
| 1058 | 3 | 0 | Nieminen, Miss. Manta Josefina | female | 29.0 | 0 | 0 | 3101297 | 7.9250 | NaN | S | NaN | NaN | NaN |
| 899 | 3 | 1 | Johnson, Mrs. Oscar W (Elisabeth Vilhelmina Berg) | female | 27.0 | 0 | 2 | 347742 | 11.1333 | NaN | S | 15 | NaN | NaN |
| 1107 | 3 | 0 | Pasic, Mr. Jakob | male | 21.0 | 0 | 0 | 315097 | 8.6625 | NaN | S | NaN | NaN | NaN |
| 832 | 3 | 0 | Goodwin, Mrs. Frederick (Augusta Tyler) | female | 43.0 | 1 | 6 | CA 2144 | 46.9000 | NaN | S | NaN | NaN | Wiltshire, England Niagara Falls, NY |
| 915 | 3 | 0 | Karlsson, Mr. Nils August | male | 22.0 | 0 | 0 | 350060 | 7.5208 | NaN | S | NaN | NaN | NaN |
| 225 | 1 | 0 | Payne, Mr. Vivian Ponsonby | male | 23.0 | 0 | 0 | 12749 | 93.5000 | B24 | S | NaN | NaN | Montreal, PQ |
| 872 | 3 | 1 | Howard, Miss. May Elizabeth | female | NaN | 0 | 0 | A. 2. 39186 | 8.0500 | NaN | S | C | NaN | NaN |
| 77 | 1 | 0 | Compton, Mr. Alexander Taylor Jr | male | 37.0 | 1 | 1 | PC 17756 | 83.1583 | E52 | C | NaN | NaN | Lakewood, NJ |
| 961 | 3 | 0 | Lennon, Miss. Mary | female | NaN | 1 | 0 | 370371 | 15.5000 | NaN | Q | NaN | NaN | NaN |
| 213 | 1 | 1 | Newell, Miss. Madeleine | female | 31.0 | 1 | 0 | 35273 | 113.2750 | D36 | C | 6 | NaN | Lexington, MA |
| 1068 | 3 | 0 | Nysveen, Mr. Johan Hansen | male | 61.0 | 0 | 0 | 345364 | 6.2375 | NaN | S | NaN | NaN | NaN |
| 317 | 1 | 1 | Williams, Mr. Richard Norris II | male | 21.0 | 0 | 1 | PC 17597 | 61.3792 | NaN | C | A | NaN | Geneva, Switzerland / Radnor, PA |
| 275 | 1 | 1 | Spedden, Mrs. Frederic Oakley (Margaretta Corn... | female | 40.0 | 1 | 1 | 16966 | 134.5000 | E34 | C | 3 | NaN | Tuxedo Park, NY |
| 840 | 3 | 0 | Haas, Miss. Aloisia | female | 24.0 | 0 | 0 | 349236 | 8.8500 | NaN | S | NaN | NaN | NaN |
| 297 | 1 | 1 | Thorne, Mrs. Gertrude Maybelle | female | NaN | 0 | 0 | PC 17585 | 79.2000 | NaN | C | D | NaN | New York, NY |
| 56 | 1 | 1 | Carter, Mr. William Ernest | male | 36.0 | 1 | 2 | 113760 | 120.0000 | B96 B98 | S | C | NaN | Bryn Mawr, PA |
| 122 | 1 | 1 | Frolicher, Miss. Hedwig Margaritha | female | 22.0 | 0 | 2 | 13568 | 49.5000 | B39 | C | 5 | NaN | Zurich, Switzerland |
| 1308 | 3 | 0 | Zimmerman, Mr. Leo | male | 29.0 | 0 | 0 | 315082 | 7.8750 | NaN | S | NaN | NaN | NaN |
| 870 | 3 | 1 | Honkanen, Miss. Eliina | female | 27.0 | 0 | 0 | STON/O2. 3101283 | 7.9250 | NaN | S | NaN | NaN | NaN |
| 108 | 1 | 1 | Fleming, Miss. Margaret | female | NaN | 0 | 0 | 17421 | 110.8833 | NaN | C | 4 | NaN | NaN |
| 787 | 3 | 0 | Eklund, Mr. Hans Linus | male | 16.0 | 0 | 0 | 347074 | 7.7750 | NaN | S | NaN | NaN | Karberg, Sweden Jerome Junction, AZ |
| 1036 | 3 | 1 | Moubarek, Mrs. George (Omine "Amenia" Alexander) | female | NaN | 0 | 2 | 2661 | 15.2458 | NaN | C | C | NaN | NaN |
| 1211 | 3 | 0 | Skoog, Mrs. William (Anna Bernhardina Karlsson) | female | 45.0 | 1 | 4 | 347088 | 27.9000 | NaN | S | NaN | NaN | NaN |
| 555 | 2 | 0 | Sedgwick, Mr. Charles Frederick Waddington | male | 25.0 | 0 | 0 | 244361 | 13.0000 | NaN | S | NaN | NaN | Liverpool |
| 695 | 3 | 0 | Burns, Miss. Mary Delia | female | 18.0 | 0 | 0 | 330963 | 7.8792 | NaN | Q | NaN | NaN | Co Sligo, Ireland New York, NY |
| 18 | 1 | 1 | Bazzani, Miss. Albina | female | 32.0 | 0 | 0 | 11813 | 76.2917 | D15 | C | 8 | NaN | NaN |
| 1125 | 3 | 0 | Petersen, Mr. Marius | male | 24.0 | 0 | 0 | 342441 | 8.0500 | NaN | S | NaN | NaN | NaN |
916 rows × 14 columns