13 Object Oriented Programming¶
13.1 Exercises¶
13.1.1 Exercise¶
Modelize a sequence with few attributes and methods
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 | class Sequence(object):
def __init__(self, identifier, comment, seq):
self.id = identifier
self.comment = comment
self.seq = self._clean(seq)
def _clean(self, seq):
"""
remove newline from the string representing the sequence
:param seq: the string to clean
:return: the string without '\n'
:rtype: string
"""
return seq.replace('\n')
def gc_percent(self):
"""
:return: the gc ratio
:rtype: float
"""
seq = self.seq.upper()
return float(seq.count('G') + seq.count('C')) / len(seq)
dna1 = Sequence('gi214', 'the first sequence', 'tcgcgcaacgtcgcctacatctcaagattca')
dna2 = Sequence('gi3421', 'the second sequence', 'gagcatgagcggaattctgcatagcgcaagaatgcggc')
|
13.1.2 Exercise¶
Instanciate 2 sequences using your Sequence class, and draw schema representing the namespaces
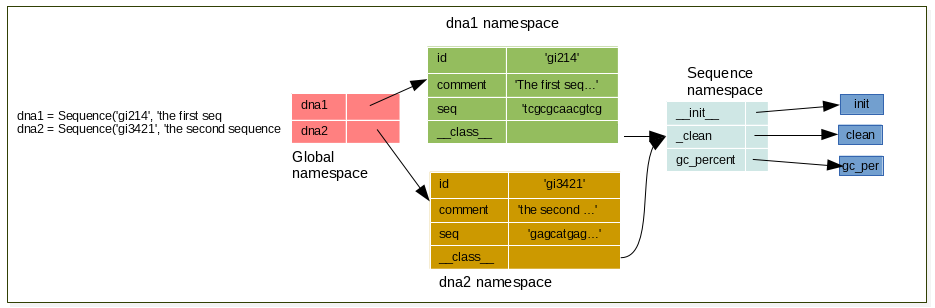

13.1.3 Exercise¶
Can you explain this result (draw namespaces to explain) ? how to modify the class variable class_attr
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 | class MyClass(object):
class_attr = 'foo'
def __init__(self, val):
self.inst_attr = val
a = MyClass(1)
b = MyClass(2)
print(a.inst_attr)
1
print(b.inst_attr)
2
print(a.class_attr == b.class_attr)
True
print(a.class_attr is b.class_attr)
True
b.class_attr = 4
print(a.class_attr)
4
del a.class_attr
MyClass.class_attr = 4
|
13.1.4 Exercise¶
Write the definition of a Point class. Objects from this class should have a
- a method show to display the coordinates of the point
- a method move to change these coordinates.
- a method dist that computes the distance between 2 points.
Note
the distance between 2 points A(x0, y0) and B(x1, y1) can be compute
(http://www.mathwarehouse.com/algebra/distance_formula/index.php)
The following python code provides an example of the expected behaviour of objects belonging to this class:
>>> p1 = Point(2, 3)
>>> p2 = Point(3, 3)
>>> p1.show()
(2, 3)
>>> p2.show()
(3, 3)
>>> p1.move(10, -10)
>>> p1.show()
(12, -7)
>>> p2.show()
(3, 3)
>>> p1.dist(p2)
1.0
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 | import math
class Point(object):
"""Class to handle point in a 2 dimensions space"""
def __init__(self, x, y):
"""
:param x: the value on the X-axis
:type x: float
:param y: the value on the Y-axis
:type y: float
"""
self.x = x
self.y = y
def show(self):
"""
:return: the coordinate of this point
:rtype: a tuple of 2 elements (float, float)
"""
return self.x, self.y
def move(self, x, y):
"""
:param x: the value to move on the X-axis
:type x: float
:param y: the value to move on the Y-axis
:type y: float
"""
self.x += x
self.y += y
def dist(self, pt):
"""
:param pt: the point to compute the distance with
:type pt: :class:`Point` object
:return: the distance between this point ant pt
:rtype: int
"""
dx = pt.x - self.x
dy = pt.y - self.y
return math.sqrt(dx ** 2 + dy ** 2)
|
point.py .
13.1.5 Exercise¶
Use OOP to modelize restriction enzyme, and sequences.
the sequence must implement the following methods
- enzyme_filter which take as a list of enzymes as argument and return a new list containing the enzymes which have binding site in sequence
the restriction enzyme must implements the following methods
- binds which take a sequence as argument and return True if the sequence contains a binding site, False otherwise.
solve the exercise 7.1.3 Exercise using this new implementation.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 |
class Sequence(object):
def __init__(self, identifier, comment, seq):
self.id = identifier
self.comment = comment
self.seq = self._clean()
def _clean(self):
"""
:param seq:
:return:
"""
return self.seq.replace('\n')
def enzyme_filter(self, enzymes):
"""
:param enzymes:
:return:
"""
enzymes_which_binds = []
for enz in enzymes:
if enz.binds(self.seq):
enzymes_which_binds.append(enz)
return
class RestrictionEnzyme(object):
def __init__(self, name, binding, cut, end, comment=''):
self._name = name
self._binding = binding
self._cut = cut
self._end = end
self._comment = comment
@property
def name(self):
return self._name
def binds(self, seq):
"""
:param seq:
:return:
"""
return self.binding in seq.seq
|
13.1.6 Exercise¶
refactor your code of 8.1.16 Exercise in OOP style programming. implements only
- size: return the number of rows, and number of columns
- get_cell: that take the number of rows, the number of columns as parameters, and returns the content of cell corresponding to row number col number
- set_cell: that take the number of rows, the number of columns as parameters, and a value and set the value val in cell specified by row number x column number
- to_str: return a string representation of the matrix
- mult: that take a scalar and return a new matrix which is the scalar product of matrix x val
you can change the name of the methods to be more pythonic
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 |
class Matrix(object):
def __init__(self, row, col, val=None):
self._row = row
self._col = col
self._matrix = []
for i in range(row):
c = [val] * col
self._matrix.append(c)
def size(self):
return self._row, self._col
def get_cell(self, row, col):
self._check_index(row, col)
return self._matrix[i][j]
def matrix_set(self, row, col, val):
self._check_index(row, col)
self._matrix[row][col] = val
def __str__(self):
s = ''
for i in range(self._row):
s += self._matrix[i]
s += '\n'
return s
def _check_index(self, row, col):
if not (0 < row <= self._row) or not (0 < col <= self._col):
raise IndexError("matrix index out of range")
|
13.1.7 Exercise¶
Use the code to read multiple sequences fasta file in procedural style and refactor it in OOP style.
use the file abcd.fasta to test your code.
What is the benefit to use oop style instead of procedural style?
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 | class Sequence(object):
def __init__(self, id_, sequence, comment=''):
self.id = id_
self.comment = comment
self.sequence = sequence
def gc_percent(self):
seq = self.sequence.upper()
return float(seq.count('G') + seq.count('C')) / float(len(seq))
class FastaParser(object):
def __init__(self, fasta_path):
self.path = fasta_path
self._file = open(fasta_path)
self._current_id = ''
self._current_comment = ''
self._current_sequence = ''
def _parse_header(self, line):
"""
parse the header line and _current_id|comment|sequence attributes
:param line: the line of header in fasta format
:type line: string
"""
header = line.split()
self._current_id = header[0][1:]
self._current_comment = ' '.join(header[1:])
self._current_sequence = ''
def __iter__(self):
return self
def next(self):
"""
:return: at each call return a new :class:`Sequence` object
:raise: StopIteration
"""
for line in self._file:
if line.startswith('>'):
# a new sequence begin
if self._current_id != '':
new_seq = Sequence(self._current_id,
self._current_sequence,
comment=self._current_comment)
self._parse_header(line)
return new_seq
else:
self._parse_header(line)
else:
self._current_sequence += line.strip()
if not self._current_id and not self._current_sequence:
self._file.close()
raise StopIteration()
else:
new_seq = Sequence(self._current_id,
self._current_sequence,
comment=self._current_comment)
self._current_id = ''
self._current_sequence = ''
return new_seq
if __name__ == '__main__':
import sys
import os.path
if len(sys.argv) != 2:
sys.exit("usage fasta_object fasta_path")
fasta_path = sys.argv[1]
if not os.path.exists(fasta_path):
sys.exit("No such file: {}".format(fasta_path))
fasta_parser = FastaParser(fasta_path)
for sequence in fasta_parser:
print("----------------")
print("{seqid} = {gc:.3%}".format(gc=sequence.gc_percent(),)
seqid = sequence.id)
|