 TC, BN, JBM, AZ
TC, BN, JBM, AZ
Institut Pasteur, Paris, 20-31 March 2017
Introductory example¶
Given a DNA sequence as an input paramter, write 4 functions to
- return a counter (dictionary) with the number of A, C, G, T letters.
- print a summary of the sequence.
- convert the sequence into its complement.
- compute the reverse complement.
# The complement function (not efficient !! there is a much better way)
def dna_complement(sequence):
complement = {'A':'T', 'C':'G', 'G':'C', 'T':'A'}
return "".join([complement[x] for x in sequence])
dna_complement('ACGTTGCA')
'TGCAACGT'
# The summary function
def summary(sequence):
txt = "sequence = %s" % sequence
txt += " length: %s\n" % str(len(sequence))
txt += "complement= %s" % dna_complement(sequence)
txt += " length: %s" % str(len(sequence))
print(txt)
summary("ACGTTGCA")
sequence = ACGTTGCA length: 8 complement= TGCAACGT length: 8
Some issues:¶
- Here above we used a simple example but with tens of parameters, handling tens of functions this may become cumbersome very quickly.
Solution: Encapsulation of the data and functions related to these data in the same structure.
- Reimplement the code above for the RNA case. This means lots of duplicated code.
Solution: inherit from a common ancestor to re-use similar code
- Data cannot be safely protected
Solution: Encapsulate the data with a protected mechanism
Class syntax: declaration and instanciation¶
We will now see how classes work and rewrite the above example using classes in Python
- A class is defined with the class keyword (instead of def for functions)
- You may see an argument named object, which is not required strictly speaking (in Python3).
# simplest version
class Sequence(object):
pass
Python2 vs Python3
In python3 all classes inherits from object so (object can be ommited). it was not the case in python2 so we had to do it explictly.Instanciation¶
- In order to create an object, we just need to call the class. We also speak of instanciation.
- The instanciation is made with the brackets
( )syntax (like a function). - Here there are no parameters but you could have, as we will see later on
s = Sequence()
Here, we have created an instance of Sequence that is stored in the variable s.
Methods¶
- A method behaves like a function but belong to a class.
- It is defined with the def keyword
- Its first argument must be consistently named self.
class DNA(object):
def example(self, param):
print(param)
def complement(self, sequence):
complement = {'A':'T', 'C':'G', 'G':'C', 'T':'A'}
return "".join([complement[x] for x in sequence])
dna = DNA()
dna.example("hello")
dna.complement("ACGT")
hello
'TGCA'
Notes:
- the function was named dna_complement. Here, the method is simply called complement (DNA is part of the class name anyway)
- Note the self in the declaration. Yet no need to use it to call the method.
- In a Python shell, thanks to the completion, we can find the methods available
Constructors¶
If we have many methods that require the sequence, it would be useful to store the sequence (encapsulate it) in the instance/object once for all. This can be done thanks to a constructor.
To provide input parameter, we need to create a constructor with a special method called __init__
class DNA(object):
def __init__(self, sequence):
self.sequence = sequence[:]
Note: here we copy the input parameter with a slice
If a positional argument is missing, methods like functions raise an exception:
dna = DNA()
--------------------------------------------------------------------------- TypeError Traceback (most recent call last) <ipython-input-46-1f2d8f7ed8a3> in <module>() ----> 1 dna = DNA() TypeError: __init__() missing 1 required positional argument: 'sequence'
Although not required, we can create a method that is called in the constructor (so only once) to pre-compute information.
from collections import Counter
class DNA(object):
def __init__(self, sequence):
self.sequence = sequence[:]
self.init()
def init(self):
self.stats = Counter(self.sequence)
dna = DNA("ACGTTT")
dna.stats
Counter({'A': 1, 'C': 1, 'G': 1, 'T': 3})
instance attribute (versus class attribute)¶
An instance attribute is a variable encapsulated in each object (each object has its own copy of the variable). Here below, sequence and length are instance attributes. So far we have seen instance attributes only.
class DNA(object):
def __init__(self, sequence):
self.sequence = sequence
self.alphabet = ['A', 'C', 'G', 'T']
def check(self):
for this in self.sequence:
if this not in self.alphaber:
raise ValueError("Found invalid letter %s" % this)
dna = DNA("ACGT")
dna.length
4
class attribute (versus instance attribute)¶
A class attribute is encapsulated in the class but is shared by all instances/objects of this class.
We can access to the class attribute from the class definition or from one of the objects.
class DNA(object):
alphabet = ['A', 'C', 'G', 'T']
def __init__(self, sequence):
self.sequence = sequence
def check(self):
for this in self.sequence:
assert this in self.alphabet
print(DNA.alphabet)
my_seq = DNA('GAATTC')
print(my_seq.alphabet)
['A', 'C', 'G', 'T'] ['A', 'C', 'G', 'T']
Since the class attribute is shared, changing the attribute has an impact on all instances
DNA.alphabet[0] = "N"
my_seq.alphabet
['N', 'C', 'G', 'T']
seq2 = DNA("ACGTAAAA")
seq2.alphabet
['N', 'C', 'G', 'T']
From functions to classes
Rewrite the first exercice using classes. Given a DNA sequence as an input parameter, we should have 4 methods to
- return a counter (dictionary) with the number of A, C, G, T letters.
- print a summary of the sequence.
- convert the sequence into its complement.
- compute the reverse complement.
class DNA:
alphabet = ['A', 'C', 'G', 'T']
complements = {'A':'T', 'C':'G', 'G':'C', 'T':'A'}
def __init__(self, sequence):
self.sequence = sequence[:]
def naive_complement(self):
new_sequence = []
for letter in self.sequence:
new_letter = self.complements[letter]
new_sequence.append(new_letter)
new_sequence = ''.join(new_sequence)
return new_sequence
def reverse_complement(self):
return self.naive_complement()[::-1]
a = DNA('ACGTGGTTGA'*1000)
%%timeit -n 10
a.naive_complement()
10 loops, best of 3: 3.29 ms per loop
A better way ?
class DNA:
alphabet = ['A', 'C', 'G', 'T']
complements = {'A':'T', 'C':'G', 'G':'C', 'T':'A'}
def __init__(self, sequence):
self.sequence = sequence[:]
def complement(self):
return "".join((self.complements[x] for x in self.sequence))
#return (self.complements[x] for x in self.sequence)
%%timeit -n 10
a = DNA("ACGTGGTTGA"*1000)
a.complement()
10 loops, best of 3: 2.37 ms per loop
A better way ?
import string
class DNA(object):
def __init__(self, sequence, complement_in=b"ACGT", complement_out=b"TGCA"):
self.sequence = sequence[:]
self._translate = bytes.maketrans(complement_in, complement_out)
def get_complement(self):
return self.sequence.translate(self._translate)
def get_reverse_complement(self):
return self.get_complement()[::-1]
def reverse(self):
return self.sequence[::-1]
dna = DNA('ACGTGGTTGA'*1000)
%timeit -n 10 dna.get_complement()
10 loops, best of 3: 7.88 µs per loop
100 times faster ! 1Gb in a second. Not bad.
Special methods¶
All classes have special methods. Special methods have two leading and two trailing underscores such as __init__ but there are many more.
class DNA(object):
def __init__(self, x):
pass
a = DNA('ACGT')
print(a)
<__main__.DNA object at 0x7fad0422e860>
When you call print(), it actually searches for __str__ method in the object definition. If not found, it looks for __repr__, which by default prints the instance type and ID. These methods can be overwriten.
The __str__ method¶
The __str__ method must return a string
class DNA(object):
def __init__(self, sequence, complement_in=b"ACGT", complement_out=b"TGCA"):
self.sequence = sequence[:]
self._translate = bytes.maketrans(complement_in, complement_out)
def complement(self):
return self.sequence.translate(self._translate)
def __str__(self):
txt = "sequence = " + self.sequence
txt += ":" + str(len(self.sequence)) + "\n"
txt += "complement= " + self.complement()
txt += ":" + str(len(self.sequence))
return txt
a = DNA('ACGT-ACGT')
print(a)
sequence = ACGT-ACGT:9 complement= TGCA-TGCA:9
The __add__ method¶
The __add__ method must return something and takes as input one parameter
class DNA(object):
def __init__(self, sequence):
self.sequence = sequence[:]
def __add__(self, other):
return self.sequence + other.sequence
def __radd_(self, other):
self.sequence = self.sequence + other.sequence
a = DNA("AAAA")
b = DNA("CCCC")
a + b
'AAAACCCC'
a += b
print(a)
AAAACCCC
'private' data¶
In Python, there is no 'private' data per se: everything is public and therefore everything can be changed (by error as well). Similarly for methods.
However, a leading underscore is conventionally added to indicate users and developers the private nature of the method or data.
class PrivateSequence(object):
def __init__(self, sequence):
self._sequence = sequence
p = PrivateSequence('acgt')
# completion does not show the variable ! but you can still access to it
# and therefore change it !
p._sequence
'acgt'
Instead of 1 underscore, you may use 2 leading underscores.
class PrivateSequence(object):
def __init__(self, sequence):
self.__sequence = sequence
p = PrivateSequence("acgt")
p.__sequence
--------------------------------------------------------------------------- AttributeError Traceback (most recent call last) <ipython-input-32-041b36a22af6> in <module>() 1 p = PrivateSequence("acgt") ----> 2 p.__sequence AttributeError: 'PrivateSequence' object has no attribute '__sequence'
p.__sequence does not exist. With 2 underscores, Python renamed it as
p._PrivateSequence__sequence
'acgt'
# But you can still rename it if you wish
p._PrivateSequence__sequence = 'A'
p._PrivateSequence__sequence
'A'
properties¶
A mechanism for checking the validity of an input and make an attribute private
The getter¶
class PrivateSequence:
def __init__(self, sequence):
self._sequence = sequence
def _get_sequence(self):
return self._sequence
sequence = property(_get_sequence)
p = PrivateSequence('acgt')
p.sequence # you can see the variable again
'acgt'
p.sequence = 'agtc' # but it is protected !
--------------------------------------------------------------------------- AttributeError Traceback (most recent call last) <ipython-input-45-45e0b9c6c617> in <module>() ----> 1 p.sequence = 'agtc' # but it is protected ! AttributeError: can't set attribute
# the same functionality but using a decorator
class PrivateSequence:
def __init__(self, sequence):
self._sequence = sequence
@property
def sequence(self):
return self._sequence
The setter¶
class PrivateSequence2:
def __init__(self, sequence):
self._sequence = sequence
def _get_sequence(self):
return self._sequence
def _set_sequence(self, sequence):
for x in sequence:
if x not in ['t','g','c','a']:
raise ValueError
self._sequence = sequence
sequence = property(_get_sequence, _set_sequence)
p = PrivateSequence2('acgt')
p.sequence = 'cgta'
print(p.sequence)
cgta
# with decorator
class PrivateSequence2:
def __init__(self, sequence):
self._sequence = sequence
@property
def sequence(self):
return self._sequence
@sequence.setter
def sequence(self, sequence):
for x in sequence:
if x not in ['t','g','c','a']:
raise ValueError
self._sequence = sequence
p = PrivateSequence2('acgt')
p.sequence = 'cgta'
print(p.sequence)
cgta
p.sequence = 'yyyy'
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) <ipython-input-27-7506d4e07f00> in <module>() 1 p = PrivateSequence2('acgt') 2 p.sequence = 'cgta' ----> 3 p.sequence = 'yyyy' <ipython-input-26-2feadb482891> in _set_sequence(self, sequence) 10 for x in sequence: 11 if x not in ['t','g','c','a']: ---> 12 raise ValueError 13 self._sequence = sequence 14 sequence = property(_get_sequence, _set_sequence) ValueError:
DNA -> RNA
Similary to the DNA class, write the RNA class
Namespace and attributes lookup¶
The LEGB rule (Local, Enclosing, Global, Built-in) still applied. But when a class is created a namespace is created. Futhermore for each instance of this a class a new namespace corresponding to this instance is created. There exists a link between the namespace of the instance and the namespace of it’s corresponding class. for example:
class Student:
school = 'Pasteur'
def __init__(self, name):
self.name = name
self.scores = []
def add_score(self, val):
self.scores.append(val)
def average(self):
av = sum(self.scores)/len(self.scores)
return av
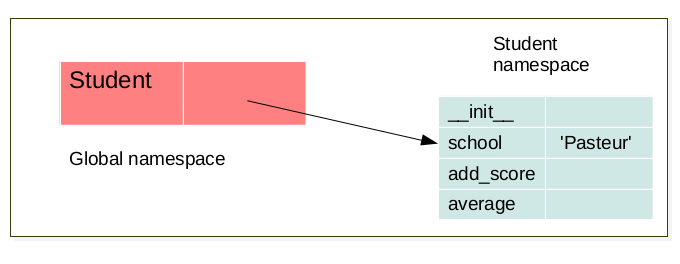
foo = Student('foo')
When an object is created, a namespace is created. This namespace is linked to its respective class namespace.
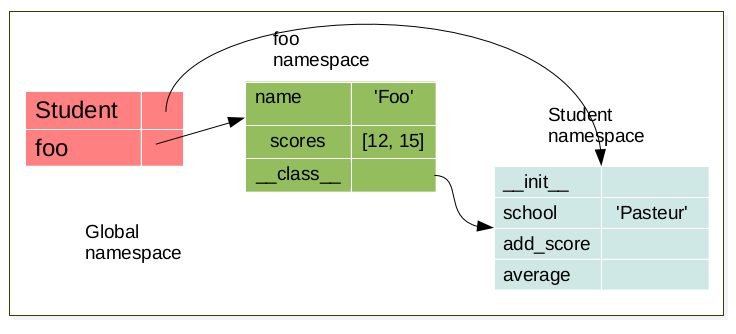
bar = Student('bar')
Each object have it’s own namespace which are linked to the class namespace.
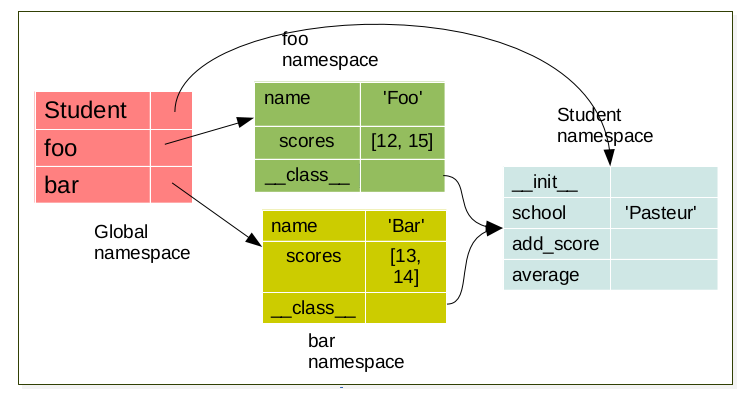
Simple Inheritance¶
So far, we have seen one main concept in object oriented programming: the encapsulation. Another powerful concept is inheritance.
In the two previous examples/ exercices, we have designed a DNA class and a RNA class. You have seen that most of the code is identical. This duplication can be avoided using inheritance.
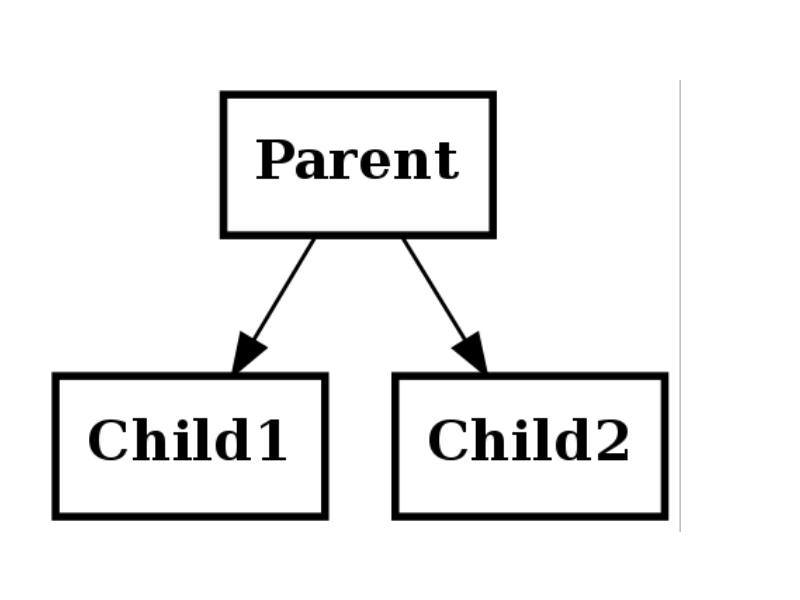
Single inheritance means a child has one parent
class DNA(object):
def __init__(self, sequence, complement_in=b"ACGT", complement_out=b"TGCA"):
self.sequence = sequence[:]
self._translate = bytes.maketrans(complement_in, complement_out)
def complement(self):
return self.sequence.translate(self._translate)
def __str__(self):
txt = "blabla"
return txt
class RNA(DNA):
def __init__(self, sequence):
super().__init__(sequence, b"ACGU", b"UGCA")
r = RNA('ACGU')
r.complement()
'UGCA'
Explanations:
- DNA class is instanciated inside RNA when the special keyword super is used
- Then, all methods found in DNA are provided to RNA
In fact RNA is not a DNA. However, RNA and DNA share in common many methods. So they should have a common parent: a Sequence. Let us rewrite the code accordingly
class Sequence(object):
def __init__(self, sequence, complement_in, complement_out):
self.sequence = sequence[:]
self._translate = bytes.maketrans(complement_in, complement_out)
def complement(self):
return self.sequence.translate(self._translate)
def __str__(self):
return "blabla"
class DNA(Sequence):
def __init__(self, sequence):
super().__init__(sequence, b"ACGT", b"TGCA")
class RNA(Sequence):
def __init__(self, sequence):
super().__init__(sequence, b"ACGU", b"UGCA")
super(__class__, self).method(arguments)
In Python 3, the old and new style are correct.
Multiple inheritance¶
Multiple inheritance means a child has several parents
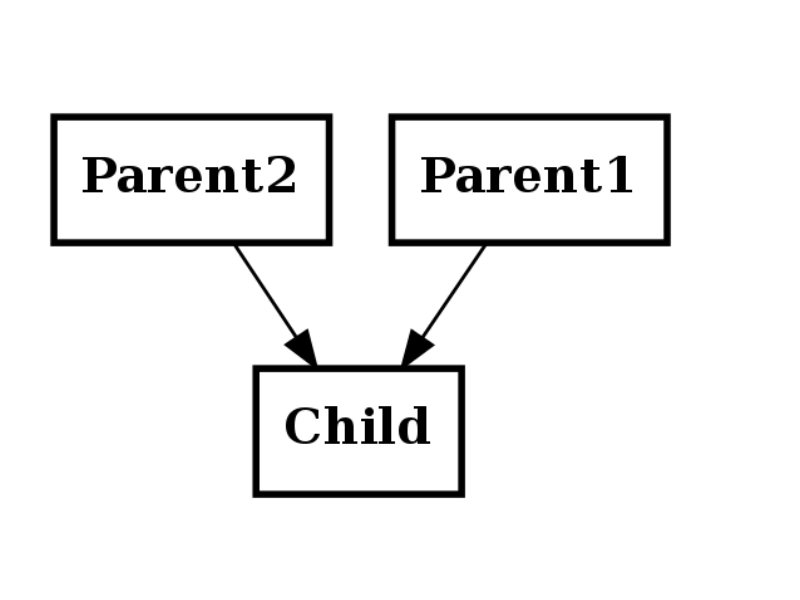
class Parent1(object):
def __init__(self):
print('initialise Parent1')
self.data_first = True
def parent1_method(self):
pass
class Parent2(object):
def __init__(self):
print('initialise Parent2')
self.data_second = True
def parent2_method(self):
pass
class Child(Parent1, Parent2):
def __init__(self):
super().__init__()
child = Child() #Note that __init__ from Parent2 is not called
initialise Parent1
Methods from the parents are exposed to the child class. If a method is found several time (same name), the priority goes from left to right (first Parent1).
child.parent1_method()
child.parent2_method()
In the previous example, only Parent1 constructor is called explicitly. There are two methods to fix the issue:
class Child2(Parent1, Parent2):
def __init__(self):
Parent1.__init__(self)
Parent2.__init__(self)
# Now, Parent1 and 2 init have been called and therefore
c = Child2()
initialise Parent1 initialise Parent2
using super() in the parents
class P1(object):
def __init__(self):
super().__init__()
print('initialise Parent1')
self.data_first = True
def test(self):
print("P1")
class P2(object):
def __init__(self):
super().__init__()
print('initialise Parent2')
self.data_second = True
def test(self):
print("P2")
class Child(P1, P2):
def __init__(self):
super().__init__()
c = Child()
c.test()
initialise Parent2 initialise Parent1 P1
Diamond inheritance¶
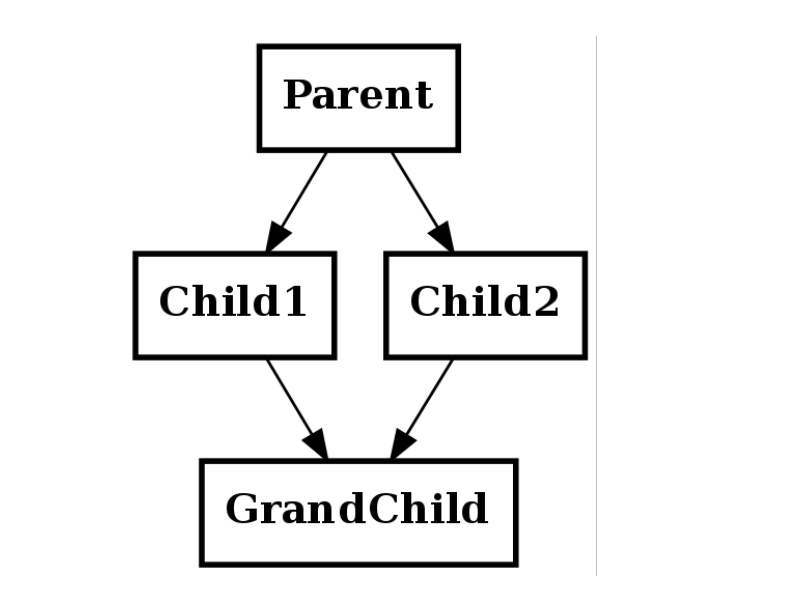
class Parent(object):
def __init__(self):
#super().__init__()
print("parent")
def hello(self):
print("hello parent")
class C1(Parent):
def __init__(self):
#super(C1, self).__init__()
print("C1")
def hello(self):
print("hello C1")
class C2(Parent):
def __init__(self):
#super().__init__()
print("C2")
def hello(self):
print("hello C2")
class GrandChild(C1, C2):
def __init__(self):
super().__init__()
print("GrandChild")
f = GrandChild()
f.hello()
#Child.mro()
C1 GrandChild hello C1
OOP programming practical session
Write the definition of a Point class. Objects from this class should have a
- a method show to display the coordinates of the point
- a method move to change these coordinates.
- a method dist that computes the distance to another point instance.
Note
the distance between 2 points A(x0, y0) and B(x1, y1) can be computed $d(AB) = \sqrt{(x_1−x_0)^2+(y_1−y_0)^2}$
Try your implementation as follows:
A = Point(1,2)
B = Point(3,4)
A.distance(B)
A.show()
A.move(3,4)
A.distance(B)
Solution¶
from math import sqrt
class Point():
def __init__(self, x, y):
self.x = x
self.y = y
def move(self, x, y):
self.x = x
self.y = y
def show(self):
print("x: {} y:{}".format(self.x, self.y))
def distance(self, other):
return sqrt((self.x - other.x)**2 + (self.y - other.y)**2)
A = Point(1,2)
B = Point(3,4)
print(A.distance(B))
A.show()
A.move(3,4)
print(A.distance(B))
2.8284271247461903 x: 1 y:2 0.0
summary:¶
A class is defined with the class keyword.
A constructor can be defined to store data or options.
Methods are defined like functions but there must be a first argument named self.
class Sequence():
def __init__(self, argument):
self.data = argument
def method1(self):
pass