11. Object Oriented Programming¶
We start this chapter by an exercise.
Try to model students and classrooms:
A student must have at least two properties: a name and scores.
A classroom must have a name and students.
Implement also:
a function that computes the average of a student’s scores;
an other one to compute the average of all students’ scores.
We can use a tuple to pack together “name” and “notes”, “name” and “students”:
1
2def student_average(student):
3 return sum(student[1]) / len(student[1])
4
5
6def class_average(classroom):
7 notes = []
8 for student in classroom[1]:
9 notes += student[1]
10 return sum(notes) / len(notes)
11
12
13if __name__ == '__main__':
14 john = ("John smith", [12, 13])
15 jim = ("Jim Morison", [13, 15, 18])
16 math = ("math", [john, jim])
17
18 print(john[0] + " average is " + str(student_average(john)))
19 print("{} average is {:2.2f}".format(jim[0], student_average(jim)))
20 print(class_average(math))
21
22 print(type(john), type(math))
But when we try to guess the type of jim and math python said tuple.
We cannot distinguish programmatically what john and math model without reading all the code.
Furthermore, the code for the average functions are not really readable.
To improve readability we can use namedtuples:
1from collections import namedtuple
2
3Student = namedtuple("Student", ("name", "scores"))
4
5
6def student_average(student):
7 return sum(student.scores) / len(student.scores)
8
9
10Classroom = namedtuple("Classroom", ("name", "students"))
11
12
13def class_average(classroom):
14 scores = []
15 for student in classroom.students:
16 scores += student.scores
17 return sum(scores) / len(scores)
18
19
20if __name__ == '__main__':
21 john = Student("John smith", [12, 13])
22 jim = Student("Jim Morison", [13, 15, 18])
23 math = Classroom("math", [john, jim])
24
25 print(john.name + " average is " + str(student_average(john)))
26 print("{} average is {:2.2f}".format(jim.name, student_average(jim)))
27 print(class_average(math))
28
29 print(type(john), type(math))
30
This solves both problems of readability and data type.
But now we want to add a new property to a student: it’s phone number. The problem is that the phone number can vary during the student’s career. Which is not allowed by the tuple.
So we can try to use a mutable data structure: a dict:
1
2
3def student_average(student):
4 return sum(student["scores"]) / len(student["scores"])
5
6
7def class_average(classroom):
8 scores = []
9 for student in classroom["students"]:
10 scores += student["scores"]
11 return sum(scores) / len(scores)
12
13
14if __name__ == '__main__':
15 john = {"name": "John smith",
16 "scores": [12, 13],
17 "phone": "0102030405"
18 }
19 jim = {"name": "Jim Morison",
20 "scores": [13, 15, 18],
21 "phone": "0506070809"
22 }
23 math = {"name": "math",
24 "students": [john, jim]
25 }
26
27 print(john["name"] + " average is " + str(student_average(john)))
28 print("{} average is {:2.2f}".format(jim["name"], student_average(jim)))
29 print(class_average(math))
30
31 print(john["phone"])
32 john["phone"] = "0101010101"
33 print(john["phone"])
34
35 print(type(john), type(math))
36
It’s not so bad but be loose the data type feature. Furthermore, we must have two different names for the average of classroom and students.
If we model student and classroom with OOP (“Object Oriented Programming”) we can pack together properties and functions that are applied on this data type:
1
2class Student:
3
4 def __init__(self, name, phone, scores=None):
5 self.name = name
6 self.phone = phone
7 if scores:
8 self.scores = scores
9 else:
10 self.scores = []
11
12
13 def average(self):
14 return sum(self.scores) / len(self.scores)
15
16
17class Classroom:
18
19 def __init__(self, name, students=None):
20 self.name = name
21 self.students = students if students else []
22
23 def average(self):
24 scores = []
25 for student in self.students:
26 scores += student.scores
27 return sum(scores) / len(scores)
28
29
30if __name__ == '__main__':
31 john = Student("John smith", "0102030405", scores=[12, 13])
32 jim = Student("Jim Morison", "0506070809", scores=[13, 15, 18])
33 math = Classroom("math", students=[john, jim])
34
35 print(john.name + " average is " + str(john.average()))
36 print("{} average is {:2.2f}.".format(jim.name, jim.average()))
37 print(f"The average of '{math.name}' class is {math.average}.")
38
39 print(john.phone)
40 john.phone = "0101010101"
41 print(john.phone)
42
43 print(type(john), type(math))
44
A class, for instance Student, is the recipe to build an object.
johnis an object, it’s an instance ofStudent.johnandjimhave the same data type, even they have distinct values.averageis called a method. It’s a function which is applied on a specific data type.
The concept of packing together data (attributes) and behavior (methods) is called encapsulation.
11.1. Concepts and Terminology¶
11.1.1. What is an Object ?¶
In programming an object is a concept. Calling elements of our programs objects is a metaphor, a useful way of thinking about them. In Python the basic elements of programming are things like strings, dictionaries, integers, functions, and so on … They are all objects. This means they have certain things in common.
In previous chapter we use the procedural style of programming. This divides your program into reusable ‘chunks’ called procedures or functions.
As much as possible you try to keep your code in these modular chunks using logic to decide which chunk is called. This makes it less mental strain to visualise what your program is doing. It also makes it easier to maintain your code. you can see which parts does what job. Improving one function (which is reused) can improve performance in several places of your program.
An object can also modeling an non real life object. For instance a parser there is no equivalent object in our lives but we need a parser to read a file in fasta format and create a sequence object so we can modeling a parser, idem with a database connection it’s not real life object but it’s very useful to think a connection as an object with properties like the port of the connection, the host of destination, … and some behaviors: connect, disconnect …
The object is very simple idea in the computing world. The objects allow us to organize code in a programs and cut things in small chunk to ease thinking about complexes ideas.
11.1.2. Classes¶
A class definition can be compared to the recipe to bake a cake. A recipe is needed to bake a cake. The main difference between a recipe (class) and a cake (an instance or an object of this class) is obvious. A cake can be eaten when it is baked, but you can’t eat a recipe, unless you like the taste of printed paper. Like baking a cake, an OOP program constructs objects according to the class definitions of the program program. A class contains variables and methods. If you bake a cake you need ingredients and instructions to bake the cake.
In Python lot of people use class, data type and type interchangeably.
To create a custom class we have to use the keyword class followed by the
name of the class the code belonging to a class in in the same block of code
(indentation):
class ClassName:
suite
class Sequence:
code ...
Some positional or keyword parameters can be add between parenthesis (these have to do with a more advanced concept in OOP: inheritance):
class ClassName(base_classes, meta=MyMetaClass):
suite
Note
PEP-8: Class names should normally use the CapWords convention.
11.1.3. Objects¶
A class is a model, a template, an object is an instance of this model. We can use the metaphor of the cake and the recipe.
You bake two cakes by following a recipe. The class is the recipe, you have two objects, the two cakes which are the instances of the same recipe.
Each cake has been made with the same ingredients but there are two independent cakes, a part of the first can be eaten whereas the second is still in the fridge:
# The model
>>> class Cake:
... pass
...
# apple_pie is an instance of Cake.
>>> apple_pie = Cake()
>>> type(apple_pie)
<class '__main__.Cake'>
# pear_pie is an instance of Cake.
>>> pear_pie = Cake()
>>> type(pear_pie)
<class '__main__.Cake'>
# The two objects are not the same.
>>> apple_pie is pear_pie
False
11.1.4. Attributes¶
Data attributes (often called simply attributes) are references to data associated to an object.
There are two kinds of attributes: instance variables, or class variables.
An instance variable is directly associated to a particular object whereas a class variable is associated to a class then all objects which are instances of this class share the same variables (to more details see section about environments). We will not encounter lot of class variables.
We can access to instance variable by its fully qualified name using the name of the instance and the name of attribute separated by a dot.
We can access to the class variables using the fully qualified name through the class or through the instances of this class.
Objects are mutable. You can change the state of an object by making an assignment to one of its attributes:
>>> class Sequence:
... # class variable
... alphabet = 'ATGC'
...
... def __init__(self, seq):
... """
... :param seq: the sequence
... :type seq: string
... """
... # instance variable
... self.sequence = seq
...
>>> ecor_1 = Sequence('GAATTC')
>>> bamh_1 = Sequence('GGATCC')
>>> print(ecor_1.sequence)
GAATTC
>>> print(bamh_1.sequence)
GGATCC
>>> print(Sequence.alphabet)
ATGC
>>> print(ecor_1.alphabet)
ATGC
>>> print(bamh_1.alphabet)
ATGC
>>> ecor_1 is bamh_1
False
>>> ecor_1.alphabet is bamh_1.alphabet
True
>>> Sequence.alphabet = 'ATGCN'
>>> print(ecor_1.alphabet)
ATGCN
11.1.5. Methods¶
In Python, methods are just attributes. They are special in the sense that they are attributes which can be executed. In Python we say callable.
A method is bound to an object. That means that this function is evaluated in the namespace of the object (see further).
1#!/usr/bin/env python3
2
3class Sequence:
4
5 # the order of the nucleotide in alphabet is important
6 # to easily compute the reverse complement
7 # class variable
8 alphabet = 'AGCT'
9
10 def __init__(self, seq):
11 """
12 :param seq: the sequence
13 :type seq: string
14 """
15 # instance variable
16 self.sequence = seq
17
18 def reverse_comp(self):
19 """
20 :return: the reverse complement of this sequence
21 :rtype: :class:`Sequence` instance
22 """
23 rev = self.sequence[::-1]
24 # Create a dict to translate nucleotides into their complement
25 # See https://docs.python.org/3/library/stdtypes.html#str.maketrans
26 table = str.maketrans(self.alphabet, self.alphabet[::-1])
27 # Use the translation dict
28 # See https://docs.python.org/3/library/stdtypes.html#str.translate
29 rev_comp = str.translate(rev, table)
30 return Sequence(rev_comp)
31
32
33my_seq = Sequence('GAATTCC')
34rev_comp = my_seq.reverse_comp()
35print(my_seq.sequence)
36print(rev_comp.sequence)
You may have notices the self parameter in function definition inside the class.
But we called the method simply as ob.func() without any arguments.
It still worked. This is because, whenever an object calls its method,
the object itself is pass as the first argument.
So, my_seq.reverse_comp() translates into Sequence.reverse_comp(my_seq).
In general, calling a method with a list of n arguments is equivalent to calling the corresponding
function with an argument list that is created by inserting the method’s object before the first argument.
For these reasons, the first argument of the function in class must be the object itself.
This is conventionally called self.
It can be named otherwise but we highly recommend to follow the convention.
11.1.6. Special methods¶
A class can implement certain operations that are invoked by special syntax (such as arithmetic operations or subscripting and slicing) by defining methods with special names. This is Python’s approach to operator overloading, allowing classes to define their own behavior with respect to language operators.
One of the biggest advantages of using Python’s magic methods is that they provide a simple way to make objects behave like built-in types. That means you can avoid ugly, counter-intuitive, and nonstandard ways of performing basic operators. In some languages, it’s common to do something like this:
if my_obj.equals(other_obj):
# do something
You could certainly do this in Python, too, but this adds confusion and is unnecessarily verbose.
Different libraries might use different names for the same operations, making the client do way more work than necessary.
With the power of magic methods, however, we can define one method (__eq__, in this case), and say what we mean instead:
if instance == other_instance:
#do something
The specials methods are defined by the language. They’re always surrounded by double underscores. There are a ton of special functions in Python.
11.1.6.1. Overloading the + Operator¶
To overload the + sign, we will need to implement the __add__ function in the class.
With great power comes great responsibility.
We can do whatever we like, inside this function.
But it is sensible to return a Point object of the coordinate sum:
class Point:
# previous definitions...
def __add__(self,other):
x = self.x + other.x
y = self.y + other.y
return Point(x,y)
Now let’s try that addition again:
>>> p1 = Point(2,3)
>>> p2 = Point(-1,2)
>>> print(p1 + p2)
(1,5)
11.1.6.2. Overloading Comparison Operators in Python¶
Python does not limit operator overloading to arithmetic operators only. We can overload comparison operators as well. Suppose, we wanted to implement the less than symbol < symbol in our Point class. Let us compare the magnitude of these points from the origin and return the result for this purpose. It can be implemented as follows:
class Point:
# previous definitions...
def __lt__(self,other):
self_mag = (self.x ** 2) + (self.y ** 2)
other_mag = (other.x ** 2) + (other.y ** 2)
return self_mag < other_mag
Some sample runs:
>>> Point(1,1) < Point(-2,-3)
True
>>> Point(1,1) < Point(0.5,-0.2)
False
>>> Point(1,1) < Point(1,1)
False
http://www.programiz.com/python-programming/operator-overloading
11.1.6.3. Comparison magic methods¶
Python provide a set of special methods to compare object: to use >, >=, ==, !=, =<, <, you have to implements the comparisons special methods (__gt__, __ge__, __eq__, __neq__, __le__, __lt__) .
- __eq__(self, other)
Defines behavior for the equality operator, ==.
- __ne__(self, other)
Defines behavior for the inequality operator, !=.
- __lt__(self, other)
Defines behavior for the less-than operator, <.
- __gt__(self, other)
Defines behavior for the greater-than operator, >.
- __le__(self, other)
Defines behavior for the less-than-or-equal-to operator, <=.
- __ge__(self, other)
Defines behavior for the greater-than-or-equal-to operator, >=.
http://www.python-course.eu/python3_magic_methods.php
11.1.6.3.1. __init__ method¶
To create an object, two steps are necessary. First a raw or uninitialized object must be created, and then the object must be initialized, ready for use. Some object-oriented languages (such as C++ and Java) combine these two steps into one, but Python keeps them separate.
When an object is created (e.g., ecor_1 = Sequence('GAATTC'),
first the special method __new__() is called to create the object,
and then the special method __init__() is called implicitly to initialize it.
In practice almost every Python class we create will require us to
reimplement only the __init__() method, since default __new__() method is al-
most always sufficient and is automatically called if we don’t provide our own
__new__() method.
Although we can create an attribute in any method, it is a good practice
to do this in the __init__ method. Thus, it is easy to know what attributes have an object
without being to read the entire code of a class:
class Sequence:
alphabet = 'ATGC'
def __init__(self, name, seq):
"""
:param seq: the sequence
:type seq: string
"""
self.name = name
self.sequence = seq
self.nucleic = True
for char in self.sequence:
if char not in self.alphabet:
self.nucleic = False
break
11.2. Namespace and attributes lookup¶
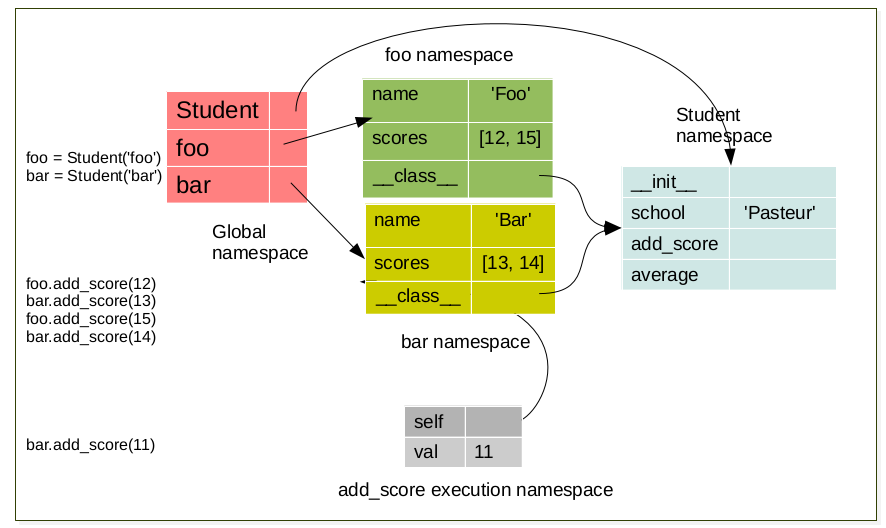
The LEGB rule (Local, Enclosing, Global, Built-in) still applied. But when a class is created a namespace is created. Furthermore, for each instance of this a class a new namespace corresponding to this instance is created. There exist a link between the namespace of the instance and the namespace of it’s corresponding class. For instance:
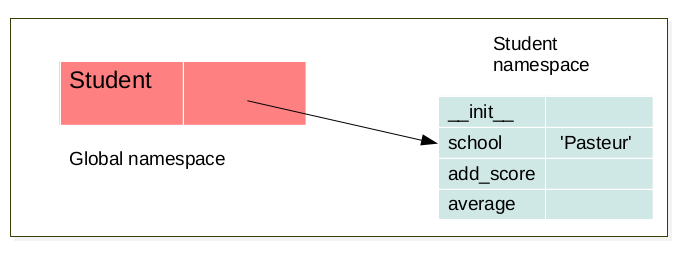
when a class is created a namespace is created.¶
class Student:
school = 'Pasteur'
def __init__(self, name):
self.name = name
self.scores = []
def add_score(self, val):
self.scores.append(val)
def average(self):
av = sum(self.scores)/len(self.scores)
return av
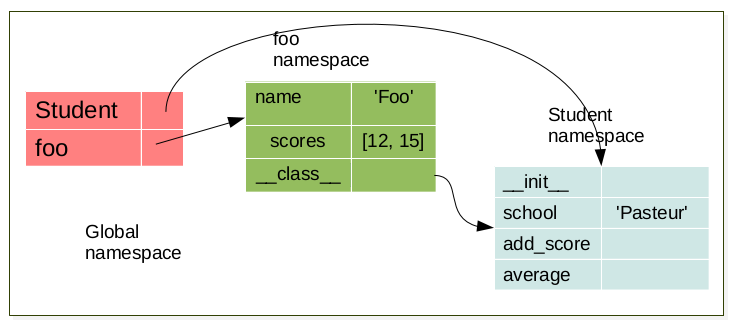
foo = Student('foo')
When an object is created, a namespace is created. This namespace is linked to its respective class namespace.
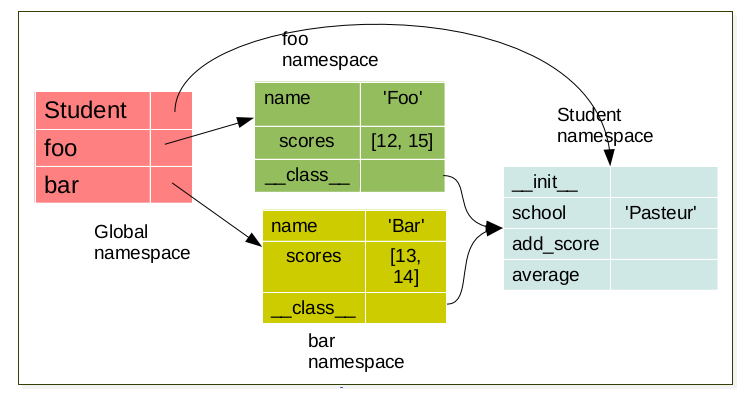
foo = Student('foo')
bar = Student('bar')
Each object have it’s own namespace which are linked to the class namespace.
foo.add_score(12)
bar.add_score(13)
foo.add_score(15)
foo.add_score(14)
bar.add_score(11)
During method execution a namespace is created which have a link to the object instance. This namespace is destroyed a the end of the method (return)
To see it in action, you can play with the code below.
1
2class TestEnv:
3
4 class_att = 1
5
6 def __init__(self):
7 self.inst_att = 2
8
9 def test(self):
10 loc_var = 3
11 print("locals:", locals())
12 print("globals:", globals())
13 print("my self:", self)
14 print("my class:", self.__class__)
15
16t = TestEnv()
17
18t.test()
11.2.1. Control the access to the attributes¶
11.2.1.1. with underscore¶
“Private” instance variables that cannot be accessed except from inside an object don’t exist in Python. However, there is a convention that is followed by most Python code: a name prefixed with an underscore (e.g. _spam) should be treated as a non-public part of the API (whether it is a function, a method or a data member). It should be considered an implementation detail and subject to change without notice.
11.3. Deleting object¶
Any attribute of an object can be deleted anytime, using the del statement.
We can even delete the object itself, using the del statement.
Actually, it is more complicated than that.
When we do s1 = Sequence('ecor1', 'GAATTC'), a new instance object is created in memory and the name s1 binds with it.
On the command del s1, this binding is removed and the name s1 is deleted from the corresponding namespace.
The object however continues to exist in memory and if no other name is bound to it, it is later automatically destroyed.
This automatic destruction of unreferenced objects in Python is also called garbage collection.
11.4. Inheritance¶
In the introduction of I mentioned that one of the objectives of OOP is to address some of the issues of software quality.
What we have seen so far, object-based programming, consists in designing programs with objects, that are built with classes. In most object-oriented programming languages, you also have a mechanism that enables classes to share code. The idea is very simple: whenever there are some commonalities between classes, why having them repeat the same code, thus leading to maintenance complexity and potential inconsistency? So the principle of this mechanism is to define some classes as being the same as other ones, with some differences.
In the example below we have design two classes to represent to kind of sequences DNA or AA. As we can see the 2 kinds of sequence have the same attributes (name and sequence) have some common methods (len, to_fasta) but have also some specific methods gc_percent and molecular_weight.
1class DNASequence(object):
2
3 alphabet = 'ATGC'
4
5 def __init__(self, name, seq):
6 """
7 :param seq: the sequence
8 :type seq: string
9 """
10 self.name = name
11 self.sequence = seq
12
13
14 def __len__(self):
15 return len(self.sequence)
16
17
18 def to_fasta(self):
19 id_ = self.name.replace(' ', '_')
20 fasta = '>{}\n'.format(id_)
21 start = 0
22 while start < len(self.sequence):
23 end = start + 80
24 fasta += self.sequence[start: end + 1] + '\n'
25 start = end
26 return fasta
27
28
29 def gc_percent(self):
30 return float(self.sequence.count('G') + self.sequence.count('C')) / len(self.sequence)
31
32
33class AASequence(object):
34
35 _water = 18.0153
36 _weight_table = {'A': 89.0932, 'C': 121.1582, 'E': 147.1293,
37 'D': 133.1027, 'G': 75.0666, 'F': 165.1891,
38 'I': 131.1729, 'H': 155.1546, 'K': 146.1876,
39 'M': 149.2113, 'L': 131.1729, 'O': 255.3134,
40 'N': 132.1179, 'Q': 146.1445, 'P': 115.1305,
41 'S': 105.0926, 'R': 174.201, 'U': 168.0532,
42 'T': 119.1192, 'W': 204.2252, 'V': 117.1463,
43 'Y': 181.1885}
44
45 def __init__(self, name, seq):
46 """
47 :param seq: the sequence
48 :type seq: string
49 """
50 self.name = name
51 self.sequence = seq
52
53
54 def __len__(self):
55 return len(self.sequence)
56
57
58 def to_fasta(self):
59 id_ = self.name.replace(' ', '_')
60 fasta = '>{}\n'.format(id_)
61 start = 0
62 while start < len(self.sequence):
63 end = start + 80
64 fasta += self.sequence[start: end + 1] + '\n'
65 start = end
66 return fasta
67
68
69 def molecular_weight(self):
70 return sum([self._weight_table[aa] for aa in self.sequence]) - (len(self.sequence) - 1) * self._water
The problem with this implementation is that a large part of code is the same in the 2 classes. It’s bad because if I have to modify a part of common code I have to do it twice. If in future I’ll need a new type of Sequence as RNA sequence I will have to duplicate code again on so on. the code will be hard to maintain. I need to keep together the common code, and be able to specify only what is specific for each type of Sequences
So we keep our two classes to deal with DNA and protein sequences, and we add a new class: Sequence, which will be a common class to deal with general sequence functions. In order to specify that a DNA (or a Protein) is a Sequence in Python is:
1class Sequence(object):
2
3 def __init__(self, name, seq):
4 """
5 :param seq: the sequence
6 :type seq: string
7 """
8 self.name = name
9 self.sequence = seq
10
11 def __len__(self):
12 return len(self.sequence)
13
14 def to_fasta(self):
15 id_ = self.name.replace(' ', '_')
16 fasta = f'>{id_}\n'
17 start = 0
18 while start < len(self.sequence):
19 end = start + 80
20 fasta += self.sequence[start: end + 1] + '\n'
21 start = end
22 return fasta
23
24
25class DNASequence(Sequence):
26
27 alphabet = 'ATGC'
28
29 def gc_percent(self):
30 return self.sequence.count('G') + self.sequence.count('C') / len(self.sequence)
31
32
33class AASequence(Sequence):
34
35 _water = 18.0153
36 _weight_table = {'A': 89.0932, 'C': 121.1582, 'E': 147.1293,
37 'D': 133.1027, 'G': 75.0666, 'F': 165.1891,
38 'I': 131.1729, 'H': 155.1546, 'K': 146.1876,
39 'M': 149.2113, 'L': 131.1729, 'O': 255.3134,
40 'N': 132.1179, 'Q': 146.1445, 'P': 115.1305,
41 'S': 105.0926, 'R': 174.201, 'U': 168.0532,
42 'T': 119.1192, 'W': 204.2252, 'V': 117.1463,
43 'Y': 181.1885}
44
45 def molecular_weight(self):
46 return sum([self._weight_table[aa] for aa in self.sequence]) - (len(self.sequence) - 1) * self._water
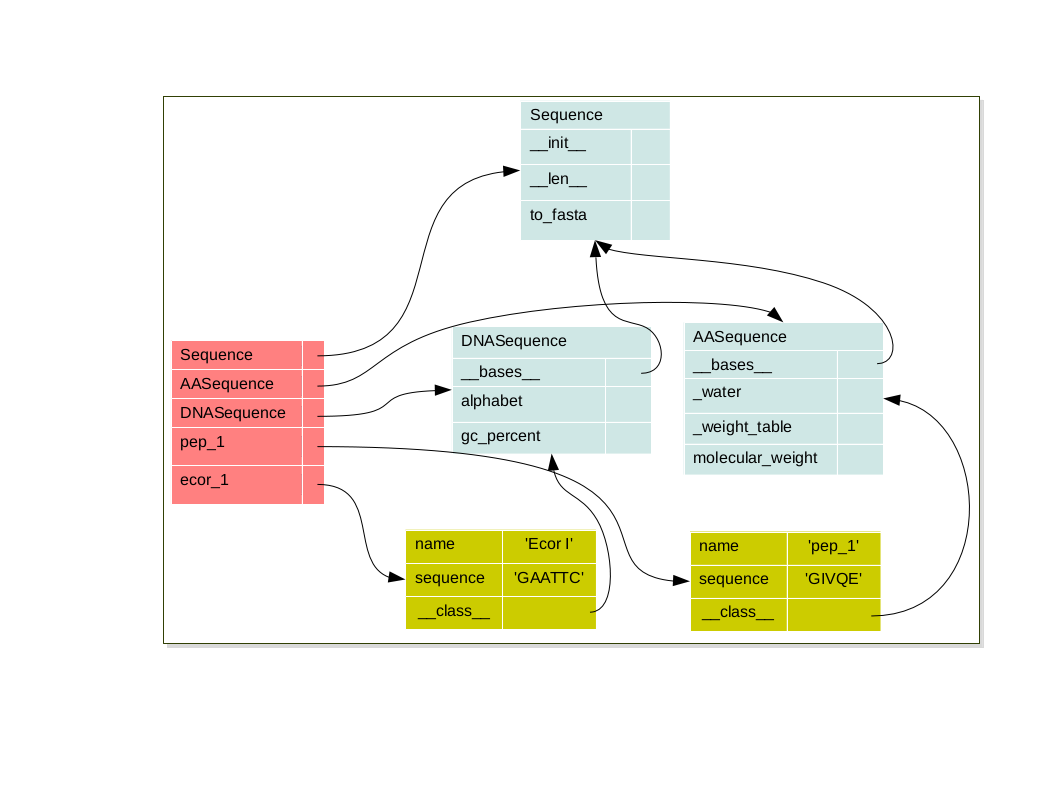

Each object have it’s own namespace which are linked to the class namespace via the special attribute:
__class__Each class is link to it’s parents namespace via the special attribute
__bases__on so on until the object class namespace
pep_1 = AASequence('pep_1', 'GIVQE')
bar = DNASequence('Ecor I', 'GAATTC')

11.4.1. Overloading¶
Overloading an attribute or a method is to redefine at a subclass level an attribute or method that exists in upper classes of a class hierarchy.
1class Sequence(object):
2
3 _water = 18.0153
4
5 def __init__(self, name, seq):
6 """
7 :param seq: the sequence
8 :type seq: string
9 """
10 self.name = name
11 self.sequence = seq
12
13 def __len__(self):
14 return len(self.sequence)
15
16 def to_fasta(self):
17 id_ = self.name.replace(' ', '_')
18 fasta = '>{}\n'.format(id_)
19 start = 0
20 while start < len(self.sequence):
21 end = start + 80
22 fasta += self.sequence[start: end + 1] + '\n'
23 start = end
24 return fasta
25
26 def molecular_weight(self):
27 return sum([self._weight_table[x] for x in self.sequence]) - (len(self.sequence) - 1) * self._water
28
29
30class DNASequence(Sequence):
31
32 alphabet = 'ATGC'
33 _weight_table = {'A': 331.2218, 'C': 307.1971, 'G': 347.2212, 'T': 322.2085}
34
35 def gc_percent(self):
36 return float(self.sequence.count('G') + self.sequence.count('C')) / len(self.sequence)
37
38 def _one_strand_molec_weight(self, seq):
39 return sum([self._weight_table[base] for base in seq]) - (len(seq) - 1) * self._water
40
41 def molecular_weight(self):
42 direct_weight = self._one_strand_molec_weight(self.sequence)
43 rev_comp = self.rev_comp()
44 rev_comp_weight = self._one_strand_molec_weight(rev_comp.sequence)
45 return direct_weight + rev_comp_weight
46
47
48class RNASequence(Sequence)
49
50 alphabet = 'AUGC'
51 _weight_table = {'A': 347.2212, 'C': 323.1965, 'G': 363.2206, 'U': 324.1813}
52
53
54class AASequence(Sequence):
55
56
57 _weight_table = {'A': 89.0932, 'C': 121.1582, 'E': 147.1293,
58 'D': 133.1027, 'G': 75.0666, 'F': 165.1891,
59 'I': 131.1729, 'H': 155.1546, 'K': 146.1876,
60 'M': 149.2113, 'L': 131.1729, 'O': 255.3134,
61 'N': 132.1179, 'Q': 146.1445, 'P': 115.1305,
62 'S': 105.0926, 'R': 174.201, 'U': 168.0532,
63 'T': 119.1192, 'W': 204.2252, 'V': 117.1463,
64 'Y': 181.1885}
65
We we overload a method sometimes we just want to add something to the parent’s method. in this case we can call
explicitly the parent’s method by using the keywords super. The syntax of this method is lit bit tricky.
the first argument must be the class that you want to retrieve the parent (usually the class you are coding),
the second argument is the object you want to retrieve the parent class (usual self) and it return a proxy to the parent
so you just have to call the method. see it in action, in the example below we overload the __init__ method
and just add 2 attribute but for the name and sequence we call the Sequence __init__ method.
1
2class Sequence(object):
3
4 _water = 18.0153
5
6 def __init__(self, name, seq):
7 """
8 :param seq: the sequence
9 :type seq: string
10 """
11 self.name = name
12 self.sequence = seq
13
14 def __len__(self):
15 return len(self.sequence)
16
17 def to_fasta(self):
18 id_ = self.name.replace(' ', '_')
19 fasta = '>{}\n'.format(id_)
20 start = 0
21 while start < len(self.sequence):
22 end = start + 80
23 fasta += self.sequence[start: end + 1] + '\n'
24 start = end
25 return fasta
26
27 def molecular_weight(self):
28 return sum([self._weight_table[x] for x in self.sequence]) - (len(self.sequence) - 1) * self._water
29
30
31class RestrictionEnzyme(Sequence):
32
33 def __init__(self, name, seq, cut, end):
34 # the line below is in python3 only
35 super().__init__(name, seq)
36 # in python2 the syntax is
37 # super(RestrictionEnzyme, self).__init__(name, seq)
38 # this syntax is also available in python3
39 self.cut = cut
40 self.end = end
41
42
43ecor1 = DNASequence('ecor I', 'GAATTC', 1, "sticky")
44print(ecor1.name, len(ecor1))
In python3 the syntax has been simplified. we can just call super() that’s all.
11.5. Polymorphism¶
The term polymorphism, in the OOP lingo, refers to the ability of an object to adapt the code to the type of the data it is processing.
Polymorphism has two major applications in an OOP language. The first is that an object may provide different implementations of one of its methods depending on the type of the input parameters. The second is that code written for a given type of data may be used on other data with another datatype as long as the other data have compatible behavior.
1def my_sum(a, b):
2 return a + b
3
4
5print("my_sum(3, 4) =", my_sum(3, 4))
6print("my_sum('three', 'four') =", my_sum('three', 'four'))
7
8
9class Sequence(object):
10
11 _water = 18.0153
12
13 def __init__(self, name, seq):
14 """
15 :param seq: the sequence
16 :type seq: string
17 """
18 self.name = name
19 self.sequence = seq
20
21 def __len__(self):
22 return len(self.sequence)
23
24 def __str__(self):
25 return f">{self.name}\n{self.sequence}"
26
27 def __add__(self, other):
28 return Sequence(f'{self.name}/{other.name}',
29 self.sequence + other.sequence)
30
31
32ecor_1 = Sequence('Ecor_I', 'GAATTC')
33bamh_1 = Sequence('Bamh I', 'GGATCC')
34
35print("my_sum(ecor_1, bamh_1) =", my_sum(ecor_1, bamh_1))
Albeit data are type, my method my_sum work equally on different type as integer, string or sequence. The my_sum method is called polymorph.
11.6. Exercises¶
Modeling a sequence with few attributes and methods
11.6.1. Exercise¶
Instantiate 2 sequences using your Sequence class, and draw a schema representing the namespaces.
11.6.2. Exercise¶
1class MyClass(object):
2
3 class_attr = 'foo'
4
5 def __init__(self, val):
6 self.inst_attr = val
7
8
9
10
11a = MyClass(1)
12b = MyClass(2)
13
14print(a.inst_attr)
15
16print(b.inst_attr)
17
18
19print(a.class_attr == b.class_attr)
20
21print(a.class_attr is b.class_attr)
22
23
24b.class_attr = 4
25
26print(a.class_attr)
27
Can you explain this result (draw namespaces to explain) ? how to modify the class variable class_attr
11.6.3. Exercise¶
Write the definition of a Point class. Objects from this class should have a
a method show to display the coordinates of the point
a method move to change these coordinates.
a method dist that computes the distance between 2 points.
Note
the distance between 2 points A(x0, y0) and B(x1, y1) can be compute
(http://www.mathwarehouse.com/algebra/distance_formula/index.php)
The following Python code provides an example of the expected behaviour of objects belonging to this class:
>>> p1 = Point(2, 3)
>>> p2 = Point(3, 3)
>>> p1.show()
(2, 3)
>>> p2.show()
(3, 3)
>>> p1.move(10, -10)
>>> p1.show()
(12, -7)
>>> p2.show()
(3, 3)
>>> p1.dist(p2)
1.0
11.6.4. Exercise¶
Use biopython to read a fasta file (sv40.fasta)
and display the following attributes:
id
name
description
seq
Use the module SeqIO from biopython.
A tutorial is available here: https://biopython.org/wiki/SeqIO
Biopython is not part of the default Python distribution. You will likely need to install it:
$ pip install biopython
Note
The “$” character is here to indicate that the above command is to be typed in a command-line interface (shell). Do not type the “$” itself.
If the above doesn’t work, you may try to install packages by forcing your specific version of Python to load pip and perform the install:
$ python3.9 -m pip install biopython
11.6.5. Exercise¶
Translate the sequence in phase 1, 2, -2
11.6.6. Exercise¶
Create a sequence with the first 42 nucleotides
Translate this sequence
Mutate the nucleotide in position 18 ‘A’ -> ‘C’
and translate the mutated sequence
see tutorial http://biopython.org/DIST/docs/tutorial/Tutorial.html#htoc28
11.6.7. Exercise¶
Open the file abcd.fasta (abcd.fasta) and convert it in genbank format.
Hint:
The seq alphabet attribute must be set to extended_protein (see the Bio.Alphabet.IUPAC module).
11.6.8. Exercice¶
Open the file abcd.fasta (abcd.fasta) and filter out sequences of lenght <= 700.
Write the results in a fasta file.
11.6.9. Exercise¶
Use OOP to model restriction enzymes and sequences.
The sequence must implement the following methods:
enzyme_filterwhich takes a list of enzymes as argument and returns a new list containing the enzymes which have binding site in the sequence
The restriction enzyme must implement the following methods:
bindswhich takes a sequence as argument and returnsTrueif the sequence contains a binding site,Falseotherwise.
Solve the exercise Exercise using this new implementation.
11.6.10. Exercise¶
Refactor your code of Exercise in OOP style programming. Implement only:
size: return the number of rows, and number of columns
get_cell: that take the number of rows, the number of columns as parameters, and returns the content of cell corresponding to row number col number
set_cell: that take the number of rows, the number of columns as parameters, and a value and set the value val in cell specified by row number x column number
to_str: return a string representation of the matrix
mult: that take a scalar and return a new matrix which is the scalar product of matrix x val
You can change the name of the methods to be more “pythonic”.
11.6.11. Exercise¶
Use the code to read multiple sequences fasta file in procedural style and refactor it in OOP style.
Use the file abcd.fasta to test your code.
What is the benefit to use OOP style instead of procedural style?